- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Re: Rube Goldberg Code
03-21-2018 10:09 AM - edited 03-21-2018 10:21 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
So I post this in the hope that someone will a) explain why my DDS code is so awful (I'm pretty sure it's because I can't use an In-Place structure, and everything is in memory, and it's chopping 20 element subsets of a 6M element array at a time), and b) what the appropriate solution is, because I'm sure it isn't what I've done. On the bright side, this works well with values of 20 and 400 and 4000 for decimation, and at 400 still appears the same in the graph for both original and decimated data. I wonder how LabVIEW's graph handles it.
The arrays are reordered in the Build Array at the end, so that the graph shows why I don't use the Decimate (cont) in Signal Operation palette. It's even less desirable if I enable averaging.

03-21-2018 11:48 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
I think you can put the "choose matching time values" for loop in the first loop? That would save building two arrays of indices. Not sure how that affects execution speed.
03-21-2018 01:50 PM - edited 03-21-2018 03:08 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
In addition to combining the two loops into one as suggested (half the parallelization overhead), I would also try replacing half the swap code with min&max. Have you tried without parallelization? (e.g. I doubt the second loop would benefit). How about combining both data into a 1D complex array inside the loop? Can you attach your code and some data so I can try a few other things?
(Maybe we should move the discussion into the LabVIEW forum. It's an optimization problem, not really Rube Goldberg (your code is too good for this :D))
03-21-2018 02:57 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
My gut instinct is that your issue is the memory allocations. I would try to profile that section of code itself and see just what is being created, specially every loop. DETT is your friend here.
Some simple changes I see include:
1) Absolutely combine the two loops, there's nothing that needs to operate on the array of indices is the individual ones beforehand. Doing so will save creation of 2 arrays of size 300k (6M/20).
2) Does it really matter if the min/max are in same order as the data? Your graph is so scrunched, that perhaps it makes sense to have 2 data arrays, one representing max, other min. There is a good chunk of overhead to do the double array build at end of each iteration of the loop.
Have you done simple timing checks to see where the "slowness" of your algorithm comes from? It could easily be items you are not thinking about like reading from TDMS, or the other 2 methods to display arrays, or even the merge signals afterward.
"I won't be wronged. I won't be insulted. I won't be laid a-hand on. I don't do these things to other people, and I require the same from them." John Bernard Books
03-21-2018 11:12 PM - edited 03-21-2018 11:13 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Thanks wiebe, Altenbach and bsvare.
As you all noted, there's no need for a second For loop. Not sure why I didn't see this...
@altenbach wrote:
In addition to combining the two loops into one as suggested (half the parallelization overhead), I would also try replacing half the swap code with min&max. Have you tried without parallelization? (e.g. I doubt the second loop would benefit). How about combining both data into a 1D complex array inside the loop?
Removing the parallelization from the loop made no noticeable difference, which surprised me, but perhaps ~300k runs isn't enough to offset the starting costs. I'll remember this when prematurely adding P in the future...
The 1D complex array is a good idea (which despite having read you suggest it in similar situations several times, didn't occur to me at all). I tried it and it 'felt' faster, but I still had a Complex to Re/Im at the end to pass back into the TDMS file (although it's connected to a False constant to a case structure, so maybe the compiler drops it).
bsvare, certainly the TDMS Read will take some time, even on an SSD, and so that might even be the main remaining element of 'slowness', but with the first (While loop) method, it was incredibly slow. After a minute or so of running, I turned on the Trace Execution option and saw that the remaining array size was still around 6M, having only done maybe around 1% of the dataset. Now, it completes in maybe 1.5s or so, which is fine for code that isn't part of anything - I just needed to decimate some data for a presentation and working in DIADEM for an hour I still hadn't gotten any further than deciding that the Resample (Stairs) or (Analog) were both pretty bad for me.
@altenbach@altenbach wrote:
Can you attach your code and some data so I can try a few other things?
(Maybe we should move the discussion into the LabVIEW forum. It's an optimization problem, not really Rube Goldberg (your code is too good for this :D))
Would you like me to? I agree this became more in-depth than I expected for a largely comic thread, but I figured there was probably some built-in node that I'd missed (beyond Max and Min, which I swapped in now). If you're curious, I've attached the VI and (reduced quantity) data to this post, but please feel free to ignore them - I hope not to need to run this code too often and just found it interesting how vastly different in performance two implementations of the same idea can be...
03-22-2018 03:19 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Maybe, but it would be a lot of work, there is a way to do it faster. The TDMS read allocates a lot of data. If you could include it in one loop it might be faster. So, read chunks to be decimated from TDMS, decimate them straight away, repeat.
I understand decimating XY graphs is not easy. What I never got is why the other graphs and charts don't decimate themselves. It puzzles me that 70MB in a graph takes seconds to render, while decimating it myself and rendering takes only .1 second. The result is exactly the same, so why isn't decimation build in?
03-22-2018 07:34 AM - edited 03-22-2018 07:35 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
@cbutcher wrote:
Removing the parallelization from the loop made no noticeable difference, which surprised me, but perhaps ~300k runs isn't enough to offset the starting costs. I'll remember this when prematurely adding P in the future...
bsvare, certainly the TDMS Read will take some time, even on an SSD, and so that might even be the main remaining element of 'slowness', but with the first (While loop) method, it was incredibly slow. After a minute or so of running, I turned on the Trace Execution option and saw that the remaining array size was still around 6M, having only done maybe around 1% of the dataset. Now, it completes in maybe 1.5s or so, which is fine for code that isn't part of anything
just found it interesting how vastly different in performance two implementations of the same idea can be...
So, a couple take aways I see from this comment set.
1) Parallelization not helping? This surprises me as the code itself doesn't have a loop to loop variability. It should have a significant boost in speed if you have more than 2 cores. There is one exception that if the loops run so fast, that parallelization overhead actually slows things down. If this is the case, I doubt the for loop itself takes any time at all.
2) First loop contains a delete from array. This is known notorious slow method. I don't remember where on the board I saw this, but I swear it's on the order of 200 to 1000x (or worse) slower than sub array the data you want. You are also resizing the array each itteration and adding and removing 70 MB chunks to RAM each round.
3)Your total time is 1.5s? Considering you have to read 70MB from disk, compute, and display in that time, I don't think that's bad at all.
All the more reason I suggest doing a timing per section. If 95% of your time is the read from tdms, it doesn't make sense to worry about optimizing the other code much more. There's anecdotal evidence this is already the case.
"I won't be wronged. I won't be insulted. I won't be laid a-hand on. I don't do these things to other people, and I require the same from them." John Bernard Books
03-22-2018 09:37 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
This thread was originally in the Rube Goldberg thread, but was suggested that it really didn't belong there. So I moved it to the regular LabVIEW forum.
03-22-2018 10:39 AM - edited 03-22-2018 10:40 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
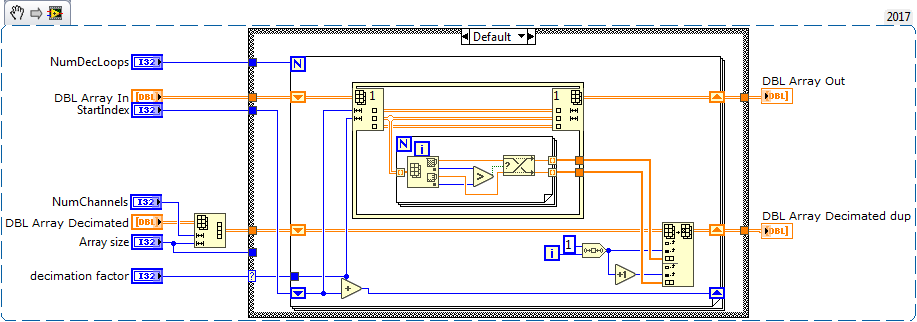
Note sure if this will help, but attached is a data decimation VI that I use in tight loops. It assumes you have a 2D data array and the data is in rows.
It operates in place, and is relatively fast although Altenbach can probably improve it by a factor of 10-100. ![]()
It decimates data based on your plot display width, so in the beginning of your program you can determine the display width and it will decimate accordingly with a min/max decimation.
The meat of the code is shown below, but the all VIs are attached in 2015 version.
Cheers,
mcduff
03-22-2018 11:46 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
So, I downloaded your code and did the basic timing analysis. Now, I don't expect this to be equal to your performance because I have a different computer and only a part of your TDMS, but worth considering for you to test.
40ms Read TDMS
03ms Parse Original Data
09ms Decimate Algorithm
03ms Graph
I changed nothing on your code other then to sequential the different major components and do a timing of each. As I see it, the Read TDMS is the 75% of time now.
"I won't be wronged. I won't be insulted. I won't be laid a-hand on. I don't do these things to other people, and I require the same from them." John Bernard Books