- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Python client to unflatten received string from TCP Labview server
Solved!10-04-2016 11:44 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Sorry, I don't understand what you mean by that. How does the byte order of the transport layer make any difference for the payload? They are completely separate.
10-04-2016 02:27 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Network byte order is generally considered a synonym to big endian byte order. Of course each protocol layer can have its own byte order, but if you talk about a specific protocol layer to be in network byte order you usually say that it is in big endian byte order. Just because the entire IP suite uses big endian byte order and calls this network byte order, doesn't mean that you can not use the term network byte order for your own higher level protocol layer.

10-04-2016 02:38 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
In terms of the message content it helps that the upper layer is in big-endian (network byte order). It keeps things consistent. Though as noted you are free to do whatever you want. From the LabVIEW side I would not reoreder the data. I would leave that up to the receiving end if necessary.
Mark Yedinak
Certified LabVIEW Architect
LabVIEW Champion
"Does anyone know where the love of God goes when the waves turn the minutes to hours?"
Wreck of the Edmund Fitzgerald - Gordon Lightfoot
10-06-2016 07:55 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
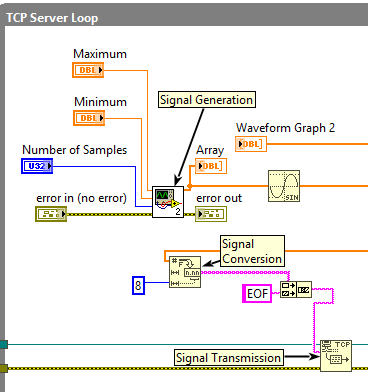
So this is how it can be done. Convert to fractional string representation. Build a protocol which has only a trailer (end of file). decode('ascii') and convert to useful representation with comma seperated values. Check the python.
@Mark_Yedinak wrote:There is no need to convert the data to ASCII before sending. You can send it in a binary form. You just need to use basic data types. You can easily send arrays of floating point or double values. You can simply flatten the data and send it. Here is an example.
For Pyhton to interpret the data the following should be used.
1D Array
First 4 bytes represent the length (number of elements) of the array. Following that are the values, each represented byte 8 bytes (double) in ANSI/IEEE Standard 754-1985 format. Single precision values would be four bytes long.
This is not true! It says in the doc: flatten 32 bit integer compliant to IEEE 754. If you decode this with python struct.unpack 'f' ('d' for 8 bit) format identifier the data is wrong. The flattening does some undocumented stuff which violates the decoding. Maybe someone mentioned the IEEE 754 to clarify numerical precision. This does not reference the actual data representation. This should be clarified.
10-06-2016 11:01 AM - edited 10-06-2016 11:58 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
I don't understand why you mark this as a solution, because it is inefficient, lossy, and clumsy and even wrong. DBLs have twice as many significant digits, and if you try to re-use that same code for signals in the microvolts, you would get significant quantization. Formatting DBL to a decimal representation then scanning it back into DBL almost never recovers the original signal bit by bit. However, it needs to transfers significantly more bytes.
Try to repeat your experiment after multiplying the sine by 1E6 or by 1E-9 and see if you still get reasonable values at the other end.
As I said, you should flatten without the size header in little endian (if on windows) and the rest will fall into place.
Currently you are not "converting", but formatting and concatenating into an ugly string without reasonable item delimiters.
10-07-2016 02:25 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Try to repeat your experiment after multiplying the sine by 1E6 or by 1E-9 and see if you still get reasonable values at the other end.
As I said, you should flatten without the size header in little endian (if on windows) and the rest will fall into place.
Currently you are not "converting", but formatting and concatenating into an ugly string without reasonable item delimiters.
I acknowledge the fact that with 8 digits after the decimal point measuring microvolts will be poor. I increased this to 12 digits after the decimal point. Even the best instruments have a noise level at 60 dBm. So submicro resolution is not really an issue. My solution does not have limitations towards higher values.
The rest you said is a claim which I frankly do not care about as long as you ignore the problem of converting these values into correct values in python. Run it try it you will see what's the problem. Nothing just falls into place. And all that chatting about endianness. To figure out the correct one you need to run the code on the receiver side TWICE. That's it.
10-07-2016 03:26 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
As I said, you should flatten without the size header in little endian (if on windows) and the rest will fall into place.
Yes in all fairness: flattening prepends size headers to everything and one can disable that primitive. In all fairness: this I did not try. Might work like that.
10-07-2016 03:27 AM - edited 10-07-2016 03:36 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
@maik.holzhey wrote:
This is not true! It says in the doc: flatten 32 bit integer compliant to IEEE 754. If you decode this with python struct.unpack 'f' ('d' for 8 bit) format identifier the data is wrong. The flattening does some undocumented stuff which violates the decoding. Maybe someone mentioned the IEEE 754 to clarify numerical precision. This does not reference the actual data representation. This should be clarified.
Well, just because you don't understand the issue at hand does not mean that Flatten does undocumented stuff. It doesn't!
But unlike most software who can not be bothered about Endianess, LabVIEW has been multiplattform since the early 1990ies and run on both Big Endian and Little Endian machines. Because of that they standardized on the Big Endian format for all flattened binary data, for part because LabVIEW originated on the Mac with the Motorola 68k CPU which was a Big Endian architecture.
If you prepend the format string for your struct.unpack() command with the > or ! character, Python will interprete the LabVIEW originated flattened binary stream correctly!
And since LabVIEW flattened data is packed too, the alignment of "none", that Python uses for these format specifiers matches up with LabVIEW too.
Here is an excerpt from the Python manual for the struct command:
Alternatively, the first character of the format string can be used to indicate the byte order, size and alignment of the packed data, according to the following table:
Character Byte order Size Alignment
| @ | native | native | native |
| = | native | standard | none |
| < | little-endian | standard | none |
| > | big-endian | standard | none |
| ! | network (= big-endian) | standard | none |
If the first character is not one of these, '@' is assumed.
If you don't specify this format specifier, then Python assumes "native", which on any Intel x86 based machine is Little Endian and therefore not what LabVIEW uses for its flattened data. In addition to that Python also uses native alignement which for all modern CPUs is usually the data element size itself up to 8 bytes.

10-07-2016 04:10 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Guys, this does not work! You can figure out endianness with two runs. You don't even have to think about it. Just run the client twice with different endianness. It is NOT the issue.
The data in python is still wrong, also disabling size headers in the flattening primitive does not work. The solution with formatting to a fractional string and the selfmade protocol is in my opinion the only solution to this.
10-07-2016 04:44 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Not having LabVIEW 2016 installed on this machine, I can't check out your code. Could you backsave it to 2015 or earlier? Or as second best option upload a snippet of your code?