- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
LabVIEW File Read Slow??
03-31-2021 03:59 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Hi,
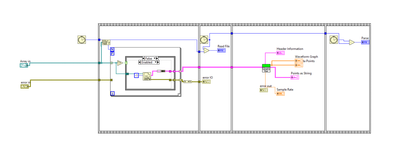
I am using the read text file VIs and it's taking upwards of 8 seconds to read these 3 CSV files into memory using the Read Text File VI....that seems way to slow...I have A LOT of these to process.
I am running LV 2020 64 bit, I have a great computer and SSD and their resource usage is less than 10%.
Here's a code snippet...
03-31-2021 04:24 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
My first reaction is to wonder why you enabled parallelism on a file access loop? Your SSD can still only pass data to the processor on the single data bus. Disable that and then see if any of those files are highly fragmented.
"Should be" isn't "Is" -Jay
03-31-2021 04:29 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Well I have some more insight here...
I can read all these files in like 70 MILLISECONDS into memory if I turn off the readline and just read text bytes...I had some colleagues read this files in Python and C++ and their file read times were similar, about 50 milliseconds or so.
So LabVIEW's readline file is just horribly slow.
My colleague also tried using LabVIEW's read text file function with a fresh copy of the file on his PC, so no fragmentation, and the difference between read lines and read characters is HUGE. Like 100mS VS 10 SECONDS huge. Wow.
Now I am wondering what the most efficient custom code I can write to split this into lines is, because taking on the order of seconds to read data from a file is not cool LabVIEW.
The disk is not slammed AT ALL. It never goes about 1% disk useage. That's why I attempted to enable parallelism.
03-31-2021 06:01 PM - edited 03-31-2021 06:06 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Are you sure you have your End of Line (EOL) properties set correctly? I don't know whether or not this would make a difference, but it does have to do with... well... end of lines.
But maybe you can avoid all that stuff by using the Read Delimited Spreadsheet.vi You can avoid all the loops and stuff by using it.
I've never noticed any difference in speed between using different read methods, and I've never heard of anyone complain about it, but like I said, maybe this can all be avoided by using the VI mentioned above.
(Mid-Level minion.)
My support system ensures that I don't look totally incompetent.
Proud to say that I've progressed beyond knowing just enough to be dangerous. I now know enough to know that I have no clue about anything at all.
Humble author of the CLAD Nugget.
03-31-2021 06:28 PM - edited 03-31-2021 06:29 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
We cannot really help unless we get some more details. What is happening in the subVI and why can't it just deal with a plain string? Why do you needs the points and the points as strings? Seems redundant!
Parsing each file (read as one single binary string) is trivial. Split off the headers (first ~105 lines) and convert the rest directly into a 1D array of SGL or DBL. What values do you actually need from the header?
Yes, parallelization is pointless here.
Reading lines is significantly more expensive that just reading a file, because every single character needs to be inspected to detect linefeeds. Then you place all lines into an array of strings with an initially unknown number of elements and each element having a random string length. You are really hammering the memory manager here. Since all files have a different number of lines, you then need to further place each it into a cluster to create a ragged 2D array. So many data copies!!! So much unnecessary work!
@xkenneth86 wrote:
Here's a code snippet...
No, that's just an image, not a snippet. A snippet has a very specific meaning in LabVIEW, because it actually contains the diagram code as metadata.
If you want help, please attach your VI and subVI. we already have some sample files.
03-31-2021 06:56 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
I decided to "see for myself". I first looked at your first .csv file. It appears to me that this is actually a pure text file, with <LF> as the end-of-line character, and no "horizontal" separators (no <comma> nor <tab> characters were noted). Thus the extension .csv is misleading -- .txt would be more appropriate.
The file is about 4.6 MB in size. It took 6.7 seconds to read this as a Text file, and resulted in a file with 600,168 lines (that's a lot!). Looking at the file, this makes sense -- almost all of the file is a list of numbers, roughly 8 bytes/line (so 8 x 600,000 = 4.8 MB).
So why does it take so long? If I change the code to read it as a string of bytes, I can read all 473464 Bytes in 10 msec. But now I have to parse it into lines, meaning search for <LF>, isolate the String, and build (one string at a time) an array of 600,000 "lines".
You Pays Your Money, You Takes Your Choice. I'm willing to wait 10 seconds and have LabVIEW do the parsing of 400+ M characters into Strings I Can Use, rather than attempt to process all of those characters, one at a time, myself.
Bob Schor
03-31-2021 07:26 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Bob,
Thanks a lot, this helps me feel a bit less crazy. I really appreciate you taking me at my word and taking your time to try it for yourself.
I hear what you're saying with the trade of between speed and convenience but it's hard for me to rationalize away LabVIEW's poor inherent performance on parsing this.
I had my colleagues who are also programmers write a bit of code to parse these files as a point of interest and they were able to produce example programs in C++ and Python that parsed these files into lists of strings in memory (reading in the file and going through the line endings) in less than 100 ms total.
So it seems like only in LabVIEW do I have to choose significantly between convenience and speed.
This code has worked well for my purposes for quite some time but I am now processing thousands of these files and so 10 seconds a set * 1000 =~ 3 hours of data processing which another language could handle in ~1.5 minutes. (10 seconds vs 100 ms.)
Don't get me wrong I am a huge fan of LabVIEW and I have built a massive backend to our production application that's written in LabVIEW.
Having to dig in and attempt to try to write some super fast byte parsing code to extract this data fast in the language I love most is kind of a bummer though.
03-31-2021 08:10 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Well, if you want to work at the "array of bytes -> array of numbers" and if you are willing to posit that there is a "header" (which you handle separately, as it is <1% of the file) and a large byte string consisting of numeric values separated by <LF>, I expect this can also be vastly speeded up. Let me give it a try and I'll get back to you ...
Bob Schor
03-31-2021 08:13 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
@xkenneth86 wrote:
I had my colleagues who are also programmers write a bit of code to parse these files as a point of interest and they were able to produce example programs in C++ and Python that parsed these files into lists of strings in memory (reading in the file and going through the line endings) in less than 100 ms total.
Interesting. Just doing my own quick testing (far from a proper benchmark), the Read Text File set to read lines took one of your files 25s. Reading the same file as pure text and then using Spreadsheet String To Array took 370ms. Parsing with Match Pattern gives me an Out of Memory error.
There are only two ways to tell somebody thanks: Kudos and Marked Solutions
Unofficial Forum Rules and Guidelines
"Not that we are sufficient in ourselves to claim anything as coming from us, but our sufficiency is from God" - 2 Corinthians 3:5
03-31-2021 08:23 PM - edited 03-31-2021 08:33 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
@xkenneth86 wrote:
Bob,
Thanks a lot, this helps me feel a bit less crazy. I really appreciate you taking me at my word and taking your time to try it for yourself.
I hear what you're saying with the trade of between speed and convenience but it's hard for me to rationalize away LabVIEW's poor inherent performance on parsing this.
I had my colleagues who are also programmers write a bit of code to parse these files as a point of interest and they were able to produce example programs in C++ and Python that parsed these files into lists of strings in memory (reading in the file and going through the line endings) in less than 100 ms total.
So it seems like only in LabVIEW do I have to choose significantly between convenience and speed.
This code has worked well for my purposes for quite some time but I am now processing thousands of these files and so 10 seconds a set * 1000 =~ 3 hours of data processing which another language could handle in ~1.5 minutes. (10 seconds vs 100 ms.)
Don't get me wrong I am a huge fan of LabVIEW and I have built a massive backend to our production application that's written in LabVIEW.
Having to dig in and attempt to try to write some super fast byte parsing code to extract this data fast in the language I love most is kind of a bummer though.
Read the whole text file and send it through split strings.vi from the advanced string functions (it used to be in hidden gems and a Forums search should turn it up) yes, the vi with the ugly, odd shaped pink icon. use /n as the delimiter. That will duplicate your text weenie coworkers efforts Leaving you with the array of strings. Inline that code, or otherwise get the debugging hooks out, and the performance will be at least on par with those other efforts.
At the risk of sounding "Blunt," the performance problem here is not with the toolbox. It's the craftsman's skill at application.
The choice between convenience and performance is also being made by your C++ & py coders. They are simply more familiar with their toolset. To be fair though, they HAVE to write the code without anyway to automaticly debug the Compiled object.
Gather the data from hardware (read from disk) .. Then process.
"Should be" isn't "Is" -Jay