- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Memory management when converting a set of PNG images into a large 3D array
Solved!09-12-2017 10:43 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Hello community,
I am working on a project that involves taking a set of binary PNG slice images from an SLA 3D printer (100+ B&W images, 1280x800 pixels per image) converting them into a 3D binary U8 array, then performing some calculations on each voxel in the array.
The program works very well for small numbers of images (smaller arrays), however, I am now trying to scale this up so that I can perform the same operation on larger 3D objects (larger 3D objects = larger 3D voxel arrays). I have already implemented some of the solutions in the Fundamentals>>Managing Performance and Memory section of the LabVIEW help. For example, I save all of my 3D arrays to the hard-drive in binary format before and after performing operations instead of keeping them in RAM, I also try not to pass the large arrays into and out of SubVIs, and I use the In Place Element Structure for performing operations wherever possible.
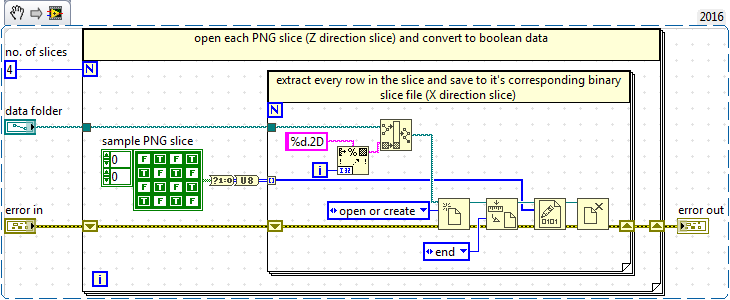
I have attached the VI that I am using to convert the PNG slices into a 3D U8 array and save this to disk. I have also attached a ZIP file of sample PNG images I am reading from, the problem occurs when I try to read very large numbers of PNG images.
I would very much appreciate any help/advice on how to proceed to scale this application up to process larger 3D arrays. The absolute upper voxel limit (taking into account the build extents of the 3D printer) is 76000x1280x800 = 7.7824E10. I know this size of 3D array is not possible so I will need to break the data into smaller chunks OR perhaps instead of converting the 2D images into a 3D array is it possible to individually address pixels in a PNG image without loading the entire image into memory?
Solved! Go to Solution.
09-13-2017 03:35 AM - edited 09-13-2017 03:38 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
The route to success is not guaranteed on this one. There will be limits and you might hit one... It would be experimental for most of us.
You've only mentioned memory as a factor, but I'm guessing execution speed is a concern as well? You'll have the trinity of execution speed\memory use\code complexity. If execution speed and memory use are given, everything must come from code complexity... If execution speed is no issue, you'll have two degrees of freedom with one fixed. It will be easier.
Here are some ideas that come to mind:
1) Can you do the calculations while getting the data from file? So no images are in memory, but are loaded when needed? This might be slow, and therefor I'd make a cache. How good this will work depends on the algorithm.
2) Since the array of bytes only contains bits, can you pack them? So there are 8 bits in a U8, instead of 1? This would reduce the data to 12.5%. Of course at the expense of code complexity.
3) There might be ways to "compress" the image data in some smart way. If there are large trunks of 1's, you might benefit from a RLE encoding or something like that. Again, at the expense of code complexity.
4) There might be ways to "compress" the 3D data in some smart way. Games use Octree's for voxel storage, and they do it for a reason. You'll use this just so it fit's in memory. Once you're ready to store to file, you can expand the data (peace by peace) as needed.
5) At the moment you are just saving the data. You might do that by loading 1 PNG, storing it, then the next. This instead of loading all PNG's and then storing them all. The data might be rotated though.
6) If you're really desperate, you can consider RAM disks. You can use this as files, but since there are (volatile) files, the are much faster. This gives more freedom in the memory department.
09-13-2017 11:43 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Thanks wiebe@CARYA for the response.
The application is not time critical so I am not too concerned about execution speed, however I would like to keep the simulation time as low as possible - but I do have some freedom here.
The calculations I need to perform are quite simple, basically I'm calculating a value for each voxel/node, the result being a 3D array of SGL floating point numbers - this is the largest array that I am using currently. I then take these values and sum them in the vertical direction from slice 0 to slice N (if that makes sense). So this is a limiting factor - I need to be able to rotate the 3D array so that I am accessing it from the horizontal direction (not the vertical direction like in the PNG slices). i.e. instead of accessing all of the X and Y voxels for a Z slice I need to be able to access all of the Z and Y voxels for an X slice - does that make sense?
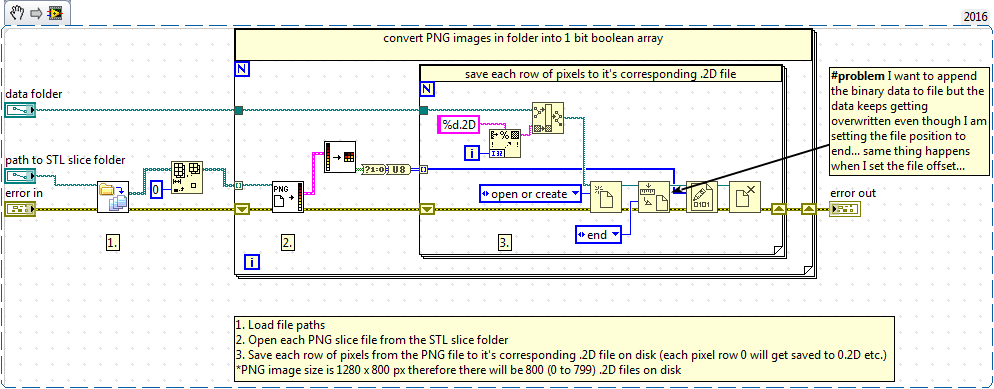
My current solution does use a cache of sorts, I store each 3D array to a folder on disk in binary format but I think the solution will be to store each array to a cache on disk in even smaller 2D chunks. However, these 2D chunks will need to be slices of the 3D array in the rotated direction (viewing from the X direction rather than the Z direction like in the PNG files). I have tried to implement this by saving each row in the PNG file to a corresponding binary file - see snippet below - but this does not work.
The binary file gets overwritten on each loop iteration leaving me with a file containing only 4 elements even though I have set the file position to the end! I've tried this approach in a few different ways but can't seem to get the new row to append to the binary file (I've tried putting the open and close file functions outside of the loop too but had the same problem) ... I'm obviously missing a trick here somewhere?
09-14-2017 02:10 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Well, the loop counter in the inner loop starts at 0 each time the outer loop increases. So you'll get 0..IL, 0..IL, 0..IL, ... OL times. Give the file a name with %d %d, so it's name has a loop counter from the outer loop, and one for the inner loop.
I'm not sure if it is efficient. I'm involved in a NDT application with 3D data and rendering, and it can be very hard to "get into it".
What I understand is that you want the totals for all pixels in one direction. So if you look at the cube from one side, sum all the pixels parallel to your line of sight. To do that, you would need a 2D buffer with the correct size. You can use a shift register to add to it as you process each .png.
But my guess is it's more complex. You'll have to think out of the box. Maybe a two-pass scheme works? First to get the total, second to calculate each pixel using the total?
09-14-2017 06:23 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
My idea with the inner loop is that it would append the rows from each new PNG to the corresponding binary file instead of overwriting it, but it's just being overwritten each time. So in reality, I would like to end up with a set of 800 2D arrays of dimension Nx1280 that contain the first row of PNG pixels from each PNG slice (0 to N)*.
I found a post on the LAVA forums (from 2006 ![]() ) that was trying to achieve the same thing and apparently the user was able to append to the binary file using the "Set file position" VI but it doesn't work in my case for some reason.
) that was trying to achieve the same thing and apparently the user was able to append to the binary file using the "Set file position" VI but it doesn't work in my case for some reason.
*each PNG is 1280x800 pixels, number of slices is N.
09-14-2017 06:39 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Try reading the file size, and put that value in Set Position. Haven't looked into it lately, but I recall that is how I usually do it (probably because using 0 and end doesn't work).
09-14-2017 08:42 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Just a thought, and I am not sure how applicable that route for your case. But whenever I see such requirements what you described, the expression "GPU computing" comes to my mind. Video cards are made to work on huge matrices in a very fast way. Ok, it also depends on how your calculations can be or not parallelized.
09-14-2017 09:57 AM - edited 09-14-2017 09:58 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
It is definitely one thing that had crossed my mind - I stumbled across the GPU toolkit while browsing the forums a while ago and it piqued my interest. However, my calculations in this case are pretty simple so I should be able to work around the memory issue (and hopefully improve speed somewhat) by splitting my 3D data into chunks.
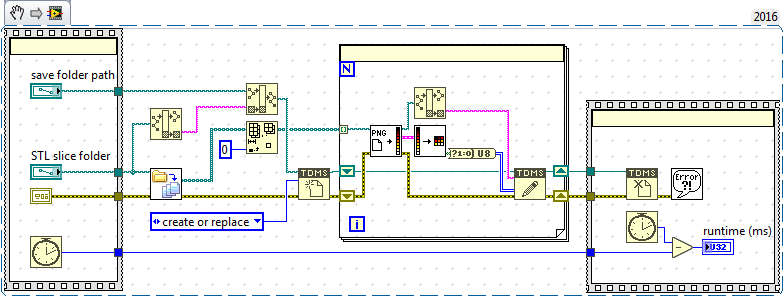
The append issue I was having I think is a misunderstanding of the write to binary format on my part. I think the TDMS format might do the trick in this case (haven't really used it much before now) but it seems that I can append my 2D slice data to different groups in the TDMS file at higher speeds than before - the code below only took 5 sec to run compared to >30 mins for a similar binary write (where I was opening and closing file references on every loop iteration).
So I think I will use the TDMS format to stream all of my array data in 2D arrays to disk, are there any caveats/recommendations for this approach? Should I use one TDMS file for all of the data or split the arrays up into different TDMS files? Is there an upper limit to TDMS file size that LabVIEW is capable of streaming to?...
09-14-2017 10:40 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
paulygon wrote: Is there an upper limit to TDMS file size that LabVIEW is capable of streaming to?...
http://digital.ni.com/public.nsf/allkb/07227F8B6D29283B86256CB1008329BC
So I guess either you move to LV 64 bitness version, or you program your code to create files below 4GBytes...
09-14-2017 12:03 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
http://digital.ni.com/public.nsf/allkb/07227F8B6D29283B86256CB1008329BC
So I guess either you move to LV 64 bitness version, or you program your code to create files below 4GBytes...
Interesting, I think I'll try installing the 64 bit version tomorrow, it might just work with the code version I am running right now...
~thanks