- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Heavy bug in "exponential fit"??
12-08-2012 04:51 AM - edited 12-08-2012 05:05 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Hi folks,
I was pretty shocked by my (hopefully wrong) finding that even the simple "exponential fit" in NI's fit-library does not behave properly, that the function does not find the minimum within acceptable tolerances.
Below I have posted the results of the NI fit compared to the result found by a standard fitting tool.
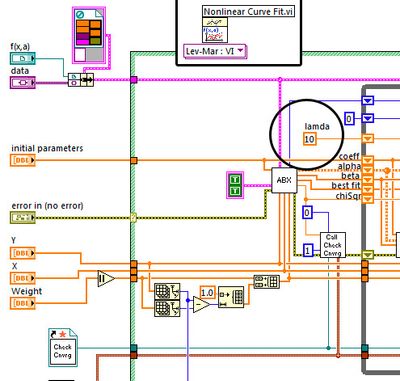
I then looked into the fit-vi and increased the number of iterations,
with modest improvent. Even setting this number to 1e8, the function only poorly found the minimum.
I knew from earlier times that the fit-functions have numerical bugs that I discovered, when trying to speed up some of these fit functions. One of the recent examples is the Levenberg Marquardt Fit, where wrong mathematics (or coding) makes the algorithm much less effective.
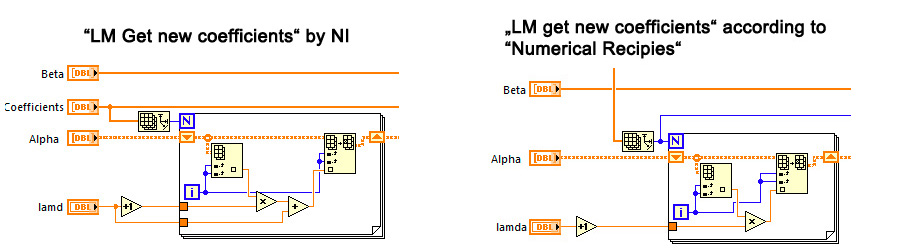
I somehow became suspicious when I realized, that the NI-LM-function starts with a lambda-value of 10 whereas in "Numerical Recipies" a start value of 0.001 is recommended.
I guess, the person who coded the function recognized, how slow his/her function was and "remedied" the error partly by increasing the start value of lambda to 10. Luckily and as the search is iterative, the function finds the minimum with acceptable precision, but is about 10 fold slower than the standard LM.
So I am asking, if somebody could confirm my findings and if I am correct, I would like to issue a general warning from using fit-functions, as I lost pretty much my trust into them.
Herbert
P.S.: I have attached a vi that does the NI-exponential fit on data stored as constants. The results of are the same with LV 2010 and LV 2012 and I expect this behaviour to be very old already.
12-08-2012 12:10 PM - edited 12-08-2012 12:24 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Thanks for raising these issues, they are important!
I just went over my copy of numerical recipes and you seem to be right with respect to the starting lambda and the formula to get the new coefficients.
I frankly never use the dedicated fit VIs (e.g. exponential fit), because I always write my own model, even in simple cases.
In any case, the Levenberg-Marquardt implementation in Nonlinear fit needs to be investigated, especially with respect to the speed of the convergence. The current nonlinear fit implementation does correctly reproduce the NIST test dataset, so the accuracy should be OK.
I implemented a test and your modifications (starting lamda, get new coeff) does speed up the NIST example quoted above.
Old algorithm: 5187 function calls
New algorithm: 3016 function calls
The results are identical!
12-08-2012 01:20 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
@Herbert wrote:
I somehow became suspicious when I realized, that the NI-LM-function starts with a lambda-value of 10 whereas in "Numerical Recipies" a start value of 0.001 is recommended.
Another small difference between the current LabVIEW implementation and the Numerical recipes book is the way lamda gets scaled:
LabVIEW: multiply by 10 or multiply by 0.4
Book: multiply by 10 or multiply by 0.1
Just wondering why the difference...
12-08-2012 01:55 PM - edited 12-08-2012 02:25 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Hi Altenbach,
as somebody from the cream of LabVIEW-experts rated my issue to be important let me give another example of poor understanding of the underlying mathematics/coding principles by the person who coded the LM-function:
In the first lesson of matlab one learns that multiplying two vectors as vectors is much much faster than multiplying the vector components in a for-loop. Second, in regression theory one learns that regression deals with overdetermined systems, means that the number of data points is larger, usually much larger than the number of parameters to fit. In my case, the number of datapoints is typically a 1000 to 10'000 times larger than the number of parameters to fit.
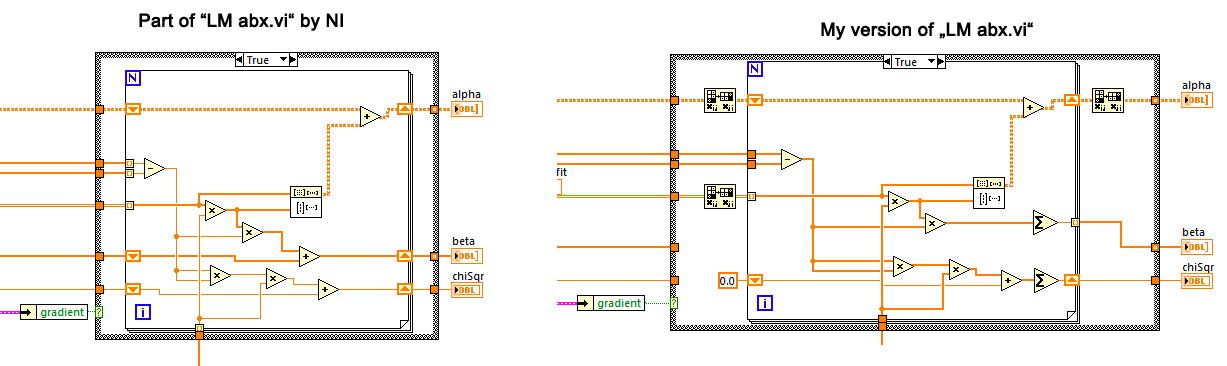
Looking at the code of NI-LM-fit it seems that it's author missed both introductory lessons as illustrated by the picture below
In order to make the wording easier let's assume that we have a model with 5 parameters and 10'000 data points:
As my picture shows, the NI-engineer did code a loop that circles 10'000 times working on vectors of the length of 5 instead of doing the reverse. When I discovered this I got another spead increase in the region of 10.
It was funny then because soon after I was scolded by a software guy in this forum to not care about basic coding principles, because code optimization functions are doing this much better than myself - I should not complain about slow compiling due to these functions that results in hopefully 20% speed increase in the executable, I should be thankful for the 20% gain by automatic code optimization as own thinking does not bring anything - a factor of 10 seems to be negligible to some people compared to 1.2?
Now my real problem is: Do I have to mistrust all mathematical functions implemented in LabVIEW or is it only the fit-functions? As I was and still am a fan of LV for more than 20 years, such discoveries really shock!
Herbert
P.S.: Please check that "My version" should illustrate the principles. I have done it in 5 minutes whereas my real implementation of this idea is a little bit different - it was specialized to the specific task and runs perfectly. So - "My version" is not tested - it might need some more tweeking.
12-08-2012 01:57 PM - edited 12-08-2012 02:13 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
@altenbach wrote:
@Herbert wrote:
I somehow became suspicious when I realized, that the NI-LM-function starts with a lambda-value of 10 whereas in "Numerical Recipies" a start value of 0.001 is recommended.
Another small difference between the current LabVIEW implementation and the Numerical recipes book is the way lamda gets scaled:
LabVIEW: multiply by 10 or multiply by 0.4
Book: multiply by 10 or multiply by 0.1
Just wondering why the difference...
Hi Altenbach,
I saw that a year ago but did not want my post to be overly long. Wanted to mention it after your response.
Greetings
Herbert
12-08-2012 03:18 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
@Herbert wrote:
In order to make the wording easier let's assume that we have a model with 5 parameters and 10'000 data points:
As my picture shows, the NI-engineer did code a loop that circles 10'000 times working on vectors of the length of 5 instead of doing the reverse. When I discovered this I got another spead increase in the region of 10.
It was funny then because soon after I was scolded by a software guy in this forum to not care about basic coding principles, because code optimization functions are doing this much better than myself - I should not complain about slow compiling due to these functions that results in hopefully 20% speed increase in the executable, I should be thankful for the 20% gain by automatic code optimization as own thinking does not bring anything - a factor of 10 seems to be negligible to some people compared to 1.2?
I'll have to study this later to see if it is correct, but just looking at the picture there are a few more optimizations possible, for example the subtraction and squaring should be moved before the loop (yes, the compiler will probably do it anyway, but why not do it directly?) There is also a "square" primitive. Newer LabVIEW versions also take much better advantage of SIMD instructions of modern processors, speeding up these primitive array operations.
I think you can also eliminate the two upper transpose operations if you swap the inputs to the outer product. Please verify!
I don't really worry about the speed of this, because it is orders of magnitude faster than my model function so it does not really matter. 😄
Can you attach your VI so I don't need to start from scratch?
12-08-2012 04:01 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Your ABX certainly does not look right. The output of the outer product (and thus alpha) now has the size of the number of points in both dimensions, but it should have the size of the number of parameters. Can you double-check?
12-08-2012 09:03 PM - edited 12-08-2012 09:04 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
@altenbach wrote:
Your ABX certainly does not look right. The output of the outer product (and thus alpha) now has the size of the number of points in both dimensions, but it should have the size of the number of parameters. Can you double-check?
sorry,
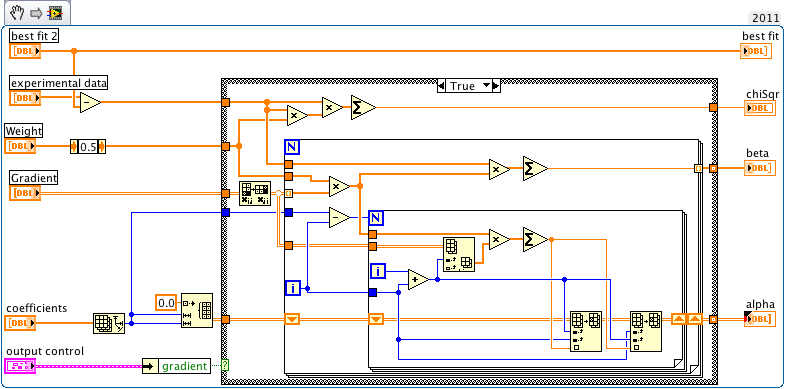
i thought it was easier but then looked up my solution from a year ago. The picture below shows the relevant part.
I agree, with the two nested for-loops it does not look as elegant as the NI-solution but - in my case - is 10 fold faster.
Herbert
12-08-2012 09:06 PM - edited 12-08-2012 09:06 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
It is silly having to look at a stamp size blurry picture. Why can't you attach the actual VI?
EDIT: OK, now it looks better 😉
12-08-2012 09:20 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Ok, I need to study it a bit more, but you seem to throw away all weights. Is that intentional?
How are you benchmarking it?