- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- « Previous
-
- 1
- 2
- Next »
Intensity graph smoothing
12-31-2015 03:01 PM - edited 12-31-2015 03:02 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
One thing is that my bilinear interpolation is relatively inefficient due to the "cute" matrix operations, making it slower than the stock solution above. However, If we use "flatter" code (See image. Similar to the inner code of the stock VIs), we can speed it up significantly because we have significantly less overhead. (For example a 10x oversampled 500x500 array takes about 10s using the stock tools, while my new code takes about 900ms (11x faster!). In addition we can parallelize the outer FOR loop for even more speed (330ms on my dual core I7 laptop (4 virtual cores due to hyperthreading), for an additional 3x speedup!). I have not tested on higher core machines, but I expect even better performance.
If course for small images and lower oversampling, things are nearly instantaeous. 😉
I would recommend to use this code. Of course your image is not square, so you need to create seperate xi and yi arrays and watch for the correct autoindexing on the two FOR loops. Should fall in place pretty quickly. 😉 Good luck.

12-31-2015 04:13 PM - edited 12-31-2015 04:15 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Of course we could wrap it all into a subVI that operates on a 2D array having one additional input specifying the smoothing factor.
Performance is pretty good.

(Typically we would just hide the x and y axes and use autoscaling, (loose fit off). Then we don't need to redo the multiplier.)
03-29-2017 10:40 AM - edited 03-29-2017 11:02 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Hello everybody!
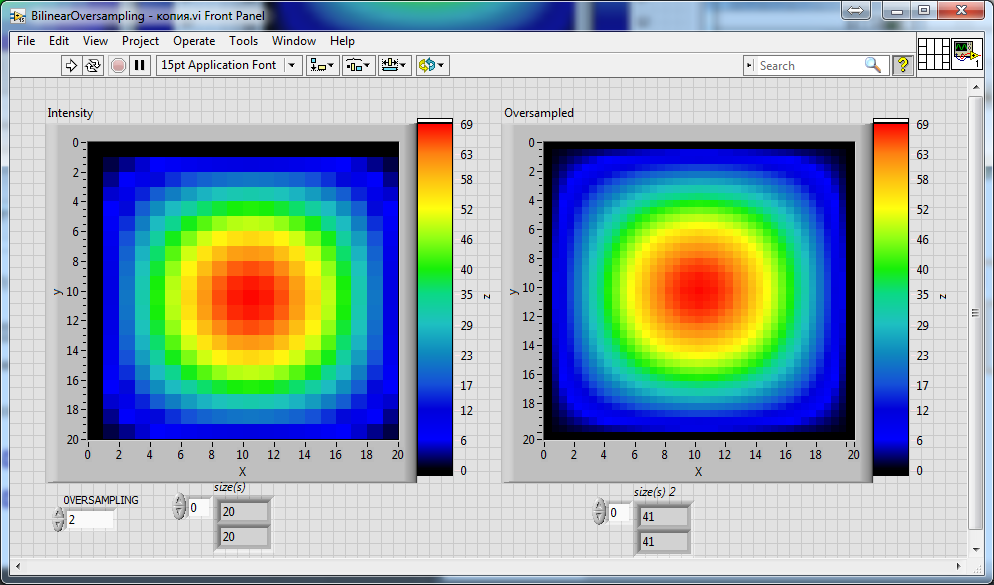
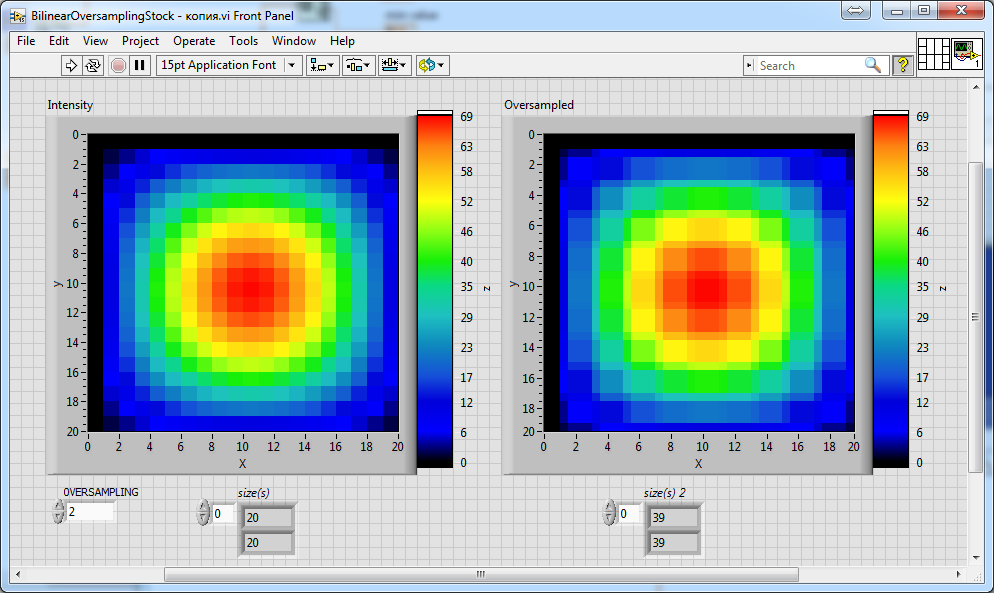
Can someone help me to improve the attached above vi's from altenbach to achieve a correct scaling factor relatively to the resolution of an input image/array? I will try to explain: if you set the scaling factor 2, you expect that the resolution of the input array or image (for example, 20x20) turns into 40x40 as the result. If you set scaling factor 3, then 60x60... And etc. But all of these attached above vi's can't do this thing. None of them... For example, I add only just 2 indicators on the front panel of each vi (attached above by altenbach), these indicators only display the size of the array before (left) and after (right) the oversampling:
So, as one can see, there is 39x39 after oversampling by the "Stock" version of the vi with scaling factor 2 (see control "OVERSAMPLING")...
And there is also 41x41 instead of 40x40 after oversampling using the vi written by altenbach with his own code.
I would be very grateful to altenbach (or anyone else) if you could help me with modifying these excellent algorithms to make them even better: to make it working more accurate in terms of resolution.
In particular, in many many applications, you need only just to increase the resolution a multiple of times (in my task the same). And the enlarged image then should be treated by another algorithms. But these algorithms very often can not handle an array, if it consists of an odd number of rows or columns etc. So, when we doing oversampling or antialiasing we often should accurately check how the vi working with output resolution.
03-31-2017 07:39 AM - edited 03-31-2017 08:07 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
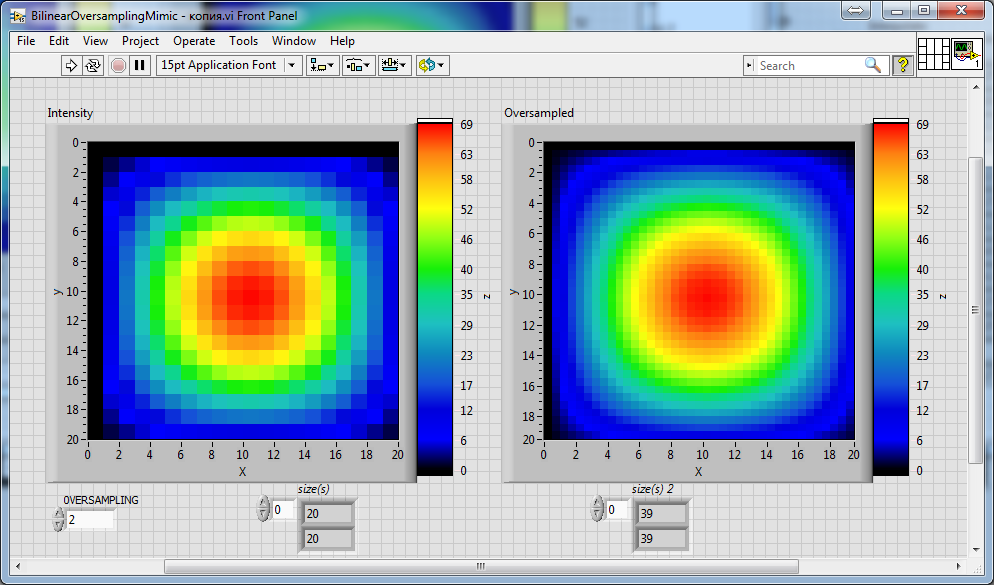
UPD. The same behavior is also observed for both the "mimic" versions of the oversamling algorithms:
The second "mimic" version (with added parallelisation) is most interesting for me (as for everyone there I think) due to it remarkable performance. So, I would be very grateful if someone helped me with the improvement of particularly this version of the oversampling algorithms. 🙂
Another question on these excellent algorithms (for which I am, of course, very grateful to altenbach with all respect for his work for us) is that they do not work correctly with the "nearest" mode of "stock" interpolation (I set it on the block-diagram):
It is obvious that problems with the Nearest mode arise precisely due to NI interpolating algorithms inaccurate work itself with the resolution of the array/image (and "Mimic" versions just copy they code). This is manifested not only in the "Nearest" mode, but also in other modes, but in them it's just not as noticeable on such a smooth test object. If we take another object, then in the modes "Cubic" or "Bilinear" it will also be more obvious:
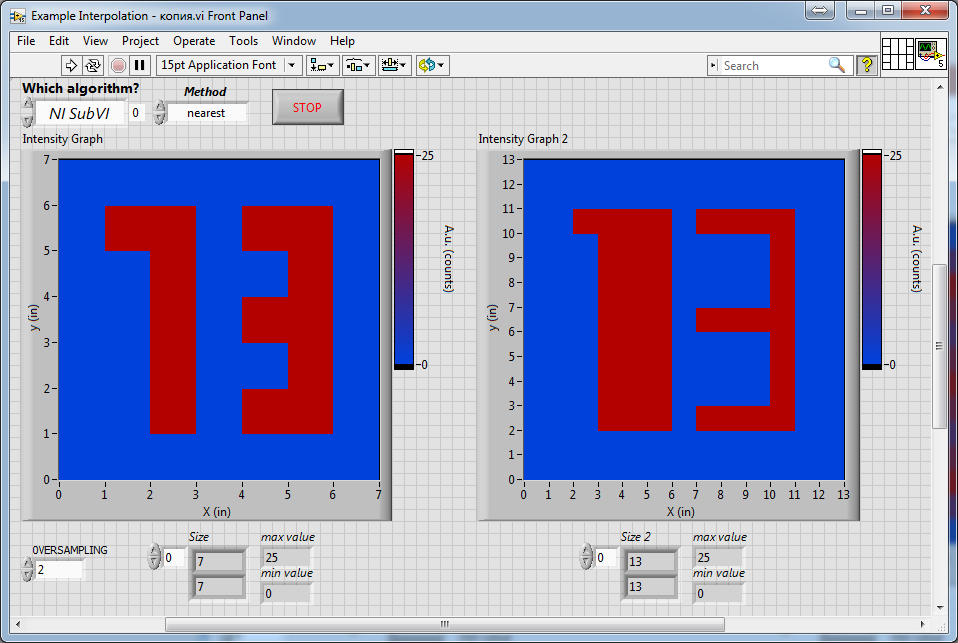


Let us compare the results presented above with the result obtained with proper accounting of the array resolution:
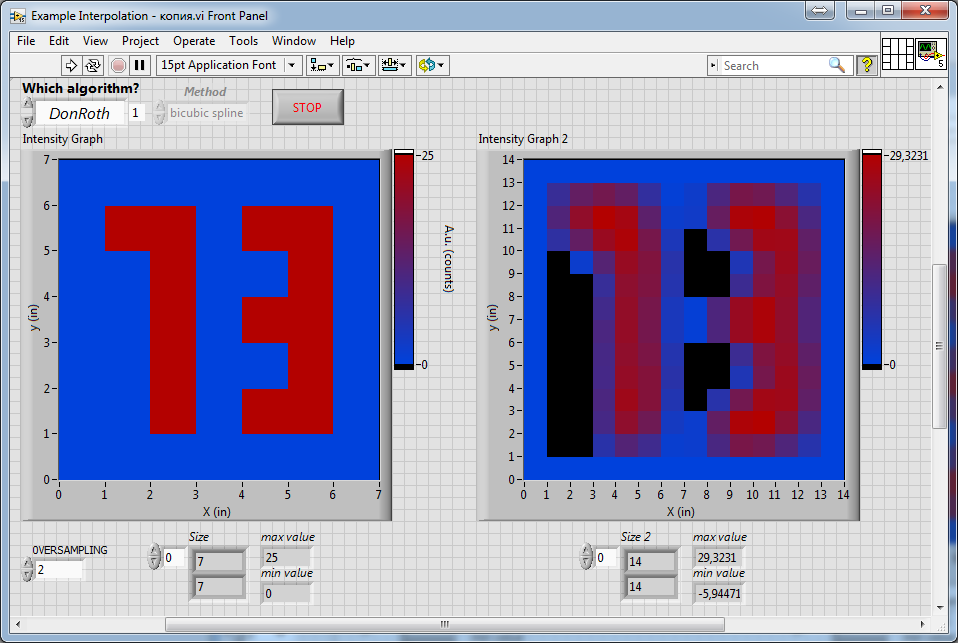 As one can see, in the last image, the thickness of the lines composing the number "3" is much closer to the original, compared to all the previous ones (which look very thin). The last result was obtained on the basis of the algorithm from DonRoth, which was attached with his post in this topic: https://forums.ni.com/t5/LabVIEW/2d-image-array-smoothing-algorithm/td-p/740816.
As one can see, in the last image, the thickness of the lines composing the number "3" is much closer to the original, compared to all the previous ones (which look very thin). The last result was obtained on the basis of the algorithm from DonRoth, which was attached with his post in this topic: https://forums.ni.com/t5/LabVIEW/2d-image-array-smoothing-algorithm/td-p/740816.Of course, I could use this subVI from DonRoth, but its performance is not as high as "Mimic" algorithm from altenbach, so on large images the conversion takes seconds, which is unacceptable for our task. In addition, in our application, the most optimal will be the use of the "Cubic" interpolation, but in the algorithm from DonRoth "Spline" method was used. Therefore, I would like somehow modify "Mimic" vi's attached by altenbach instead of using this vi from DonRoth.
The Block Diagram of the VI used to obtain all these results (presented within this spoiler):
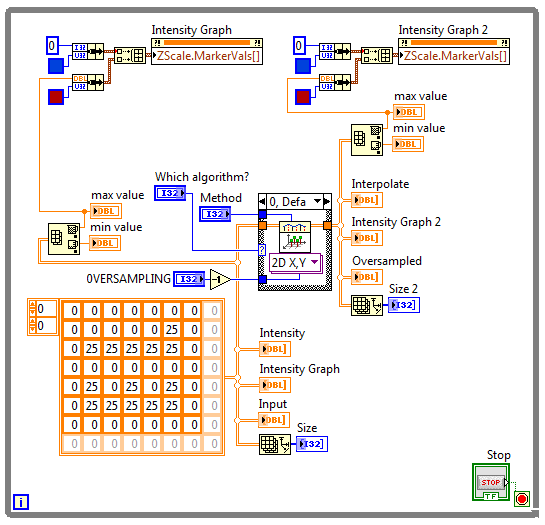 So, I just used here a stock "interpolate 2D" SubVI from NI palette.
So, I just used here a stock "interpolate 2D" SubVI from NI palette.P.S.: I apologize for overposting, I just did not have time to write everything yesterday. I have to spend a lot of time translating from my native language to English. 😃 And yesterday there was simply not enough free time to indicate all the necessary details in the message.
03-31-2017 08:07 AM - edited 03-31-2017 08:10 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Hi nik,
they do not work correctly with the "nearest" mode of "stock" interpolation
The do work correctly!
They do exactly what you told them to do: to pick the nearest value…
Each interpolation style uses its own "assumptions" on the data - the assumptions could be true or false. It's up to the programmer to choose the "correct" style…
Can someone help me to improve the attached above vi's from altenbach to achieve a correct scaling factor relatively to the resolution of an input image/array?
Well, I would never start to improve something provided by Christian! 😄
The solution to your question is quite simple: you need to set the Ramp function to produce a ramp with the "correct" number of steps…
03-31-2017 08:18 AM - edited 03-31-2017 08:31 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Thanks for the comment! 🙂 I understand you perfectly. Of course, these algorithms were developed precisely within the framework of such approach/approximation, and they working well in its boundaries (it is also required for certain tasks, I understand it). I just do not try to write about it all in my posts completely precisely in terms of scientific terminology (and other such things). I just indicated my task by simplified way (as much precise as I could to do it, because English is not my native language =). So, from such point of view, the essence of the matter does not change for me, since my question was completely practical: how do I make these algorithms work exactly as I described above? (I did not find any suitable parameters in this stock NI subVI's).
The solution to your question is quite simple: you need to set the Ramp function to produce a ramp with the "correct" number of steps…
Thank you very much! I'm just not very experienced in LabView, so even such simple things may not be clear to me. 🙂
But, I already was trying to modify it (to delete "Decrenement"), but the vi's not working with such change:
03-31-2017 08:28 AM - edited 03-31-2017 08:31 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
Hi nik,
It is obvious that problems with the Nearest mode arise precisely due to NI interpolating algorithms inaccurate work itself with the resolution of the array/image
In which way are they working "inaccurate"?
They work exactly as is written in the LabVIEW help…
how do I make these algorithms work exactly as I described above?
How do you want them to behave? (You only told what is disturbing you.)
When you need an algorithm with YOUR own requirements you need to program this algorithm.
The scaling problem from your first post is easy to solve as mentioned above.
03-31-2017 08:43 AM - edited 03-31-2017 08:46 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
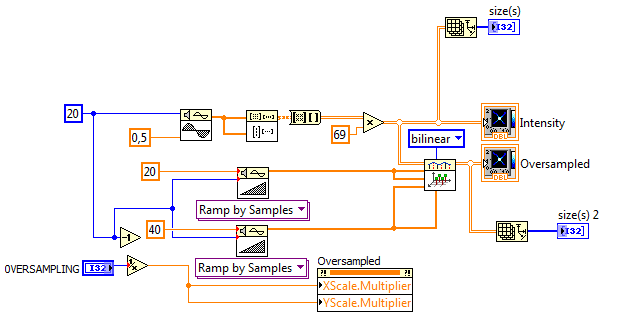
Thank you, again!... And I apologize for the stupid questions etc... 🙂 I am just figured out with help of your advise, how I need to modify this code:
 Finally:
Finally: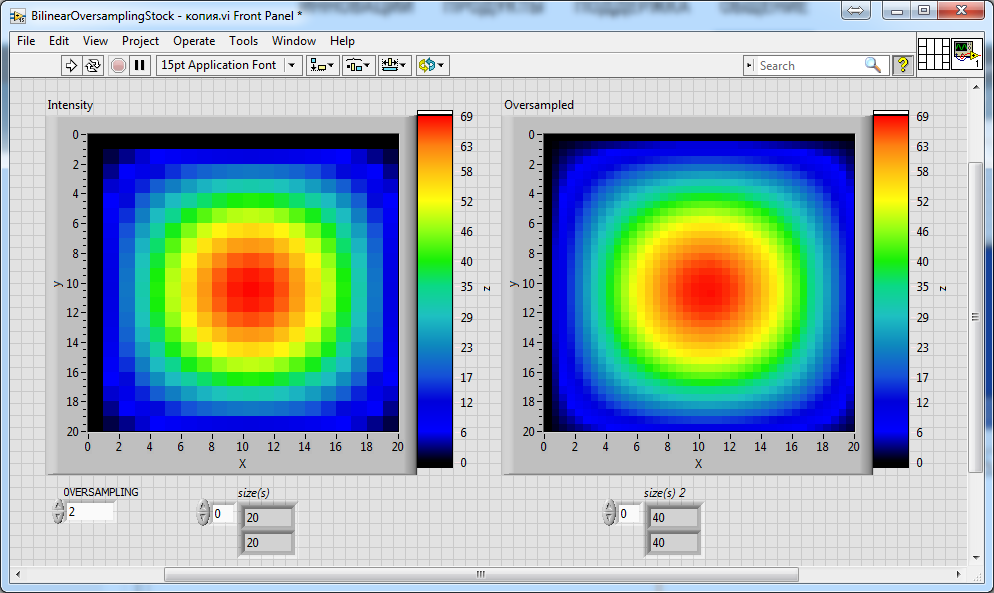
06-01-2020 10:52 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report to a Moderator
First of all, your image is not square, so you need to create ramps for X, Y, Xi, Yi independently based on the dimension sizes (minus one). It makes no sense to subtract 1 from the 2D array data. I assume you know how to get the array size of the image.
Do you have the 256bit colormap used for your Z scale?
Your image is quite large. What final size do you want after resampling? Do you understand the purpose of the various ramps?
- « Previous
-
- 1
- 2
- Next »